Counting Chromosomes, Examples
I was recently asked to look at some ES cell chromosomes by Colleen Karlo in the Alzheimer's center. I thought this was a useful example of some problems that can be encountered with counting chromosomes. Here is the commentary on the pictures:
First some general impressions. A lot of the spreads are too small to count. What normally happens is the first couple of tries are rather lackluster, and then it becomes much easier with repetition. What I look for are well separated spreads: 1-2, 4-4, 5-4 are good examples. Generally the resolution using crude karyotyping is rather poor, so I don't put much stock in morphology, rather I concentrate on getting accurate numbers of chromosomes. Anything below 40 chromosomes is either nonviable or a splatter-punk and is therefore ignored. Anything over 40, especially 41 or 42 is a concern. When I get one of these I recount it and check in the same field for additional 41/2s. In my hands, when a line goes aneuploid these 41/2s quickly take over and this is very obvious on the slides--consistent 41/2s in every valid spread in every field. From counting your chromosomes I got a mixture: a few 40s and a few 42s (see below for breakdown). Unfortunately it's just not enough good spreads to tell.
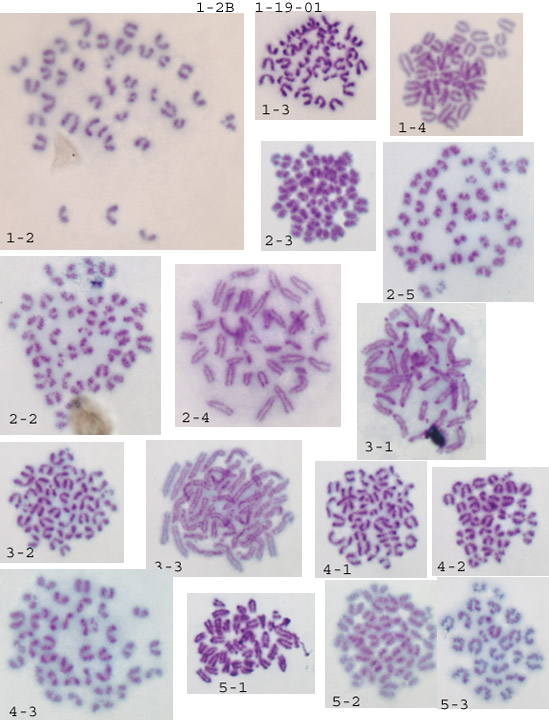
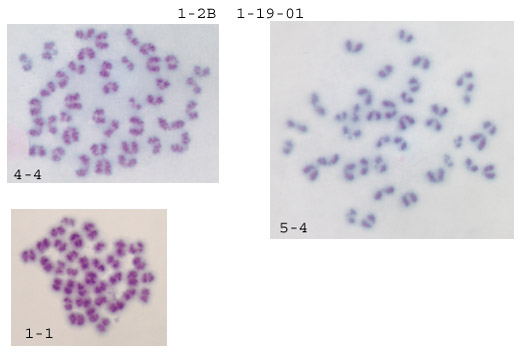
My standard procedure is to scan at low magnification for well separated spreads within a field, then flip to an oil immersion high magnification lense to count. Anything below 40 is ignored. When I get a confident 40 I move to another field. Counting 4-5 fields per slide for 4-5 slides will usually generate at least 20 confident counts. If the line has gone 41/2 it's obvious.
Now the specific counts and scoring:
|
Field |
#chromosomes |
score |
|
1-2 |
38 |
punk (ignore) |
|
2-5 |
42 |
aneuploid |
|
2-2 |
42 |
aneuploid |
|
2-4 |
40 |
euploid |
|
4-3 |
40 |
euploid |
|
1-4 |
too small |
punk |
|
4-2 |
too small |
punk |
|
5-3 |
too small |
punk |
|
4-4 |
40 |
euploid |
|
5-4 |
38 |
punk |
|
1-1 |
36 |
punk |
Assuming the naming is slide#-field#, then it looks like all the aneuploids were in two different fields on slide 2. The euploids were on 3 different fields on 2 different slides. As I said before, just not enough of a sample to tell. It does look like the procedure for making the spreads is working fine. It'll probably just come down to more hours logged on the microscope (sorry).
<< previous | next >>